8 changed files with 1325 additions and 414 deletions
Split View
Diff Options
-
+2 -0.gitignore
-
+531 -414covid-model.ipynb
-
+791 -0covid-model.md
-
BINoutput_11_1.png
-
BINoutput_18_1.png
-
BINoutput_4_1.png
-
BINoutput_7_1.png
-
+1 -0requirements.txt
+ 2
- 0
.gitignore
View File
| @ -0,0 +1,2 @@ | |||
| covid/ | |||
| .ipynb_checkpoints/ | |||
+ 531
- 414
covid-model.ipynb
File diff suppressed because it is too large
View File
+ 791
- 0
covid-model.md
View File
| @ -0,0 +1,791 @@ | |||
| ```python | |||
| # Reading data | |||
| import os | |||
| import git | |||
| import shutil | |||
| import tempfile | |||
| # Create temporary dir | |||
| t = tempfile.mkdtemp() | |||
| d = 'lwc/topics/covid19/covid-model' | |||
| # Clone into temporary dir | |||
| git.Repo.clone_from('http://gmarx.jumpingcrab.com:8088/COVID-19/covid19-data.git', | |||
| t, branch='master', depth=1) | |||
| # Delete files | |||
| #os.remove('README.txt') | |||
| shutil.rmtree('data') | |||
| #shutil.rmtree('secondTest') | |||
| # Copy desired file from temporary dir | |||
| shutil.move(os.path.join(t, 'data'), '.') | |||
| # Remove temporary dir | |||
| shutil.rmtree(t) | |||
| ``` | |||
| ```python | |||
| import pandas as pd | |||
| import numpy as np | |||
| import os | |||
| def loadData(path, file): | |||
| csvPath=os.path.join(path, file) | |||
| return pd.read_csv(csvPath) | |||
| ``` | |||
| ```python | |||
| # import jtplot submodule from jupyterthemes | |||
| from jupyterthemes import jtplot | |||
| PATH=os.path.join("data") | |||
| covid_data=loadData(PATH,"time-series-19-covid-combined.csv") | |||
| covid_data.head() | |||
| ``` | |||
| <div> | |||
| <style scoped> | |||
| .dataframe tbody tr th:only-of-type { | |||
| vertical-align: middle; | |||
| } | |||
| .dataframe tbody tr th { | |||
| vertical-align: top; | |||
| } | |||
| .dataframe thead th { | |||
| text-align: right; | |||
| } | |||
| </style> | |||
| <table border="1" class="dataframe"> | |||
| <thead> | |||
| <tr style="text-align: right;"> | |||
| <th></th> | |||
| <th>Date</th> | |||
| <th>Country/Region</th> | |||
| <th>Province/State</th> | |||
| <th>Lat</th> | |||
| <th>Long</th> | |||
| <th>Confirmed</th> | |||
| <th>Recovered</th> | |||
| <th>Deaths</th> | |||
| </tr> | |||
| </thead> | |||
| <tbody> | |||
| <tr> | |||
| <th>0</th> | |||
| <td>2020-01-22</td> | |||
| <td>Afghanistan</td> | |||
| <td>NaN</td> | |||
| <td>33.0</td> | |||
| <td>65.0</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>1</th> | |||
| <td>2020-01-23</td> | |||
| <td>Afghanistan</td> | |||
| <td>NaN</td> | |||
| <td>33.0</td> | |||
| <td>65.0</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>2</th> | |||
| <td>2020-01-24</td> | |||
| <td>Afghanistan</td> | |||
| <td>NaN</td> | |||
| <td>33.0</td> | |||
| <td>65.0</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>3</th> | |||
| <td>2020-01-25</td> | |||
| <td>Afghanistan</td> | |||
| <td>NaN</td> | |||
| <td>33.0</td> | |||
| <td>65.0</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>4</th> | |||
| <td>2020-01-26</td> | |||
| <td>Afghanistan</td> | |||
| <td>NaN</td> | |||
| <td>33.0</td> | |||
| <td>65.0</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| </tbody> | |||
| </table> | |||
| </div> | |||
| ```python | |||
| from sklearn.model_selection import train_test_split | |||
| train_set, test_set=train_test_split(covid_data,test_size=0.2,random_state=42) | |||
| train_cp=train_set.copy() | |||
| ``` | |||
| ```python | |||
| %matplotlib inline | |||
| covid_data.hist() | |||
| ``` | |||
| array([[<matplotlib.axes._subplots.AxesSubplot object at 0x11e46ca50>, | |||
| <matplotlib.axes._subplots.AxesSubplot object at 0x116caea90>], | |||
| [<matplotlib.axes._subplots.AxesSubplot object at 0x11e65dd10>, | |||
| <matplotlib.axes._subplots.AxesSubplot object at 0x11e6a16d0>], | |||
| [<matplotlib.axes._subplots.AxesSubplot object at 0x11e6d3ed0>, | |||
| <matplotlib.axes._subplots.AxesSubplot object at 0x11e716710>]], | |||
| dtype=object) | |||
|  | |||
| ```python | |||
| %matplotlib inline | |||
| import matplotlib.pyplot as plt | |||
| covid_mexico = covid_data[covid_data['Country/Region']=='Mexico'] | |||
| covid_mexico.shape | |||
| ``` | |||
| (63, 8) | |||
| ```python | |||
| ``` | |||
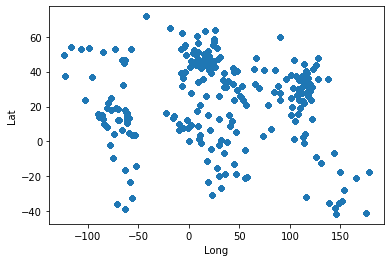
| ```python | |||
| covid_data.plot(kind="scatter", x="Long", y="Lat") | |||
| ``` | |||
| <matplotlib.axes._subplots.AxesSubplot at 0x12b7c9910> | |||
|  | |||
| ```python | |||
| from datetime import datetime | |||
| #covid_mexico['Date'] =pd.to_datetime(covid_mexico.Date, format="%Y-%m-%d") | |||
| mexico_sort=covid_mexico.sort_values(by='Date', ascending=True) | |||
| mexico_sort | |||
| ``` | |||
| <div> | |||
| <style scoped> | |||
| .dataframe tbody tr th:only-of-type { | |||
| vertical-align: middle; | |||
| } | |||
| .dataframe tbody tr th { | |||
| vertical-align: top; | |||
| } | |||
| .dataframe thead th { | |||
| text-align: right; | |||
| } | |||
| </style> | |||
| <table border="1" class="dataframe"> | |||
| <thead> | |||
| <tr style="text-align: right;"> | |||
| <th></th> | |||
| <th>Date</th> | |||
| <th>Country/Region</th> | |||
| <th>Province/State</th> | |||
| <th>Lat</th> | |||
| <th>Long</th> | |||
| <th>Confirmed</th> | |||
| <th>Recovered</th> | |||
| <th>Deaths</th> | |||
| </tr> | |||
| </thead> | |||
| <tbody> | |||
| <tr> | |||
| <th>9954</th> | |||
| <td>2020-01-22</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9955</th> | |||
| <td>2020-01-23</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9956</th> | |||
| <td>2020-01-24</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9957</th> | |||
| <td>2020-01-25</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9958</th> | |||
| <td>2020-01-26</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>0</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>...</th> | |||
| <td>...</td> | |||
| <td>...</td> | |||
| <td>...</td> | |||
| <td>...</td> | |||
| <td>...</td> | |||
| <td>...</td> | |||
| <td>...</td> | |||
| <td>...</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10012</th> | |||
| <td>2020-03-20</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>164</td> | |||
| <td>4.0</td> | |||
| <td>1</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10013</th> | |||
| <td>2020-03-21</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>203</td> | |||
| <td>4.0</td> | |||
| <td>2</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10014</th> | |||
| <td>2020-03-22</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>251</td> | |||
| <td>4.0</td> | |||
| <td>2</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10015</th> | |||
| <td>2020-03-23</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>316</td> | |||
| <td>4.0</td> | |||
| <td>3</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10016</th> | |||
| <td>2020-03-24</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>367</td> | |||
| <td>NaN</td> | |||
| <td>4</td> | |||
| </tr> | |||
| </tbody> | |||
| </table> | |||
| <p>63 rows × 8 columns</p> | |||
| </div> | |||
| ```python | |||
| mexico_filter = mexico_sort[mexico_sort['Confirmed']!=0] | |||
| mexico_filter | |||
| ``` | |||
| <div> | |||
| <style scoped> | |||
| .dataframe tbody tr th:only-of-type { | |||
| vertical-align: middle; | |||
| } | |||
| .dataframe tbody tr th { | |||
| vertical-align: top; | |||
| } | |||
| .dataframe thead th { | |||
| text-align: right; | |||
| } | |||
| </style> | |||
| <table border="1" class="dataframe"> | |||
| <thead> | |||
| <tr style="text-align: right;"> | |||
| <th></th> | |||
| <th>Date</th> | |||
| <th>Country/Region</th> | |||
| <th>Province/State</th> | |||
| <th>Lat</th> | |||
| <th>Long</th> | |||
| <th>Confirmed</th> | |||
| <th>Recovered</th> | |||
| <th>Deaths</th> | |||
| </tr> | |||
| </thead> | |||
| <tbody> | |||
| <tr> | |||
| <th>9991</th> | |||
| <td>2020-02-28</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>1</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9992</th> | |||
| <td>2020-02-29</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>4</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9993</th> | |||
| <td>2020-03-01</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>5</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9994</th> | |||
| <td>2020-03-02</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>5</td> | |||
| <td>0.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9995</th> | |||
| <td>2020-03-03</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>5</td> | |||
| <td>1.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9996</th> | |||
| <td>2020-03-04</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>5</td> | |||
| <td>1.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9997</th> | |||
| <td>2020-03-05</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>5</td> | |||
| <td>1.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9998</th> | |||
| <td>2020-03-06</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>6</td> | |||
| <td>1.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>9999</th> | |||
| <td>2020-03-07</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>6</td> | |||
| <td>1.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10000</th> | |||
| <td>2020-03-08</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>7</td> | |||
| <td>1.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10001</th> | |||
| <td>2020-03-09</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>7</td> | |||
| <td>1.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10002</th> | |||
| <td>2020-03-10</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>7</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10003</th> | |||
| <td>2020-03-11</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>8</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10004</th> | |||
| <td>2020-03-12</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>12</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10005</th> | |||
| <td>2020-03-13</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>12</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10006</th> | |||
| <td>2020-03-14</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>26</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10007</th> | |||
| <td>2020-03-15</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>41</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10008</th> | |||
| <td>2020-03-16</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>53</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10009</th> | |||
| <td>2020-03-17</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>82</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10010</th> | |||
| <td>2020-03-18</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>93</td> | |||
| <td>4.0</td> | |||
| <td>0</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10011</th> | |||
| <td>2020-03-19</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>118</td> | |||
| <td>4.0</td> | |||
| <td>1</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10012</th> | |||
| <td>2020-03-20</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>164</td> | |||
| <td>4.0</td> | |||
| <td>1</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10013</th> | |||
| <td>2020-03-21</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>203</td> | |||
| <td>4.0</td> | |||
| <td>2</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10014</th> | |||
| <td>2020-03-22</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>251</td> | |||
| <td>4.0</td> | |||
| <td>2</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10015</th> | |||
| <td>2020-03-23</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>316</td> | |||
| <td>4.0</td> | |||
| <td>3</td> | |||
| </tr> | |||
| <tr> | |||
| <th>10016</th> | |||
| <td>2020-03-24</td> | |||
| <td>Mexico</td> | |||
| <td>NaN</td> | |||
| <td>23.6345</td> | |||
| <td>-102.5528</td> | |||
| <td>367</td> | |||
| <td>NaN</td> | |||
| <td>4</td> | |||
| </tr> | |||
| </tbody> | |||
| </table> | |||
| </div> | |||
| ```python | |||
| n=mexico_filter.shape[0] | |||
| days=np.arange(1,n+1,1) | |||
| days | |||
| ``` | |||
| array([ 1, 2, 3, 4, 5, 6, 7, 8, 9, 10, 11, 12, 13, 14, 15, 16, 17, | |||
| 18, 19, 20, 21, 22, 23, 24, 25, 26]) | |||
| ```python | |||
| #mexico_filter = mexico_sort[mexico_sort['Confirmed']!=0] | |||
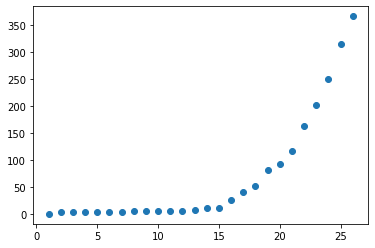
| plt.scatter(x=days, y=mexico_filter['Confirmed']) | |||
| ``` | |||
| <matplotlib.collections.PathCollection at 0x12acc8290> | |||
|  | |||
| ```python | |||
| ``` | |||
| ```python | |||
| from scipy.optimize import curve_fit | |||
| def exponential(x, a,k, b): | |||
| return a*np.exp(x*k) + b | |||
| potp, pcov = curve_fit(exponential, days, mexico_filter['Confirmed']) | |||
| potp | |||
| ``` | |||
| array([ 1.07768657, 0.22640743, -3.90363561]) | |||
| ```python | |||
| ``` | |||
| ```python | |||
| ``` | |||
| ```python | |||
| ``` | |||
| ```python | |||
| ``` | |||
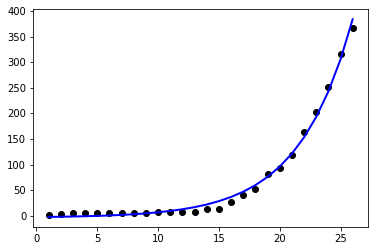
| ```python | |||
| # Plot outputs | |||
| plt.scatter(days, mexico_filter['Confirmed'], color='black') | |||
| plt.plot(days,exponential(days,*potp), color='blue', linewidth=2) | |||
| ``` | |||
| [<matplotlib.lines.Line2D at 0x12b888fd0>] | |||
|  | |||
BIN
output_11_1.png
View File
BIN
output_18_1.png
View File
BIN
output_4_1.png
View File
BIN
output_7_1.png
View File
+ 1
- 0
requirements.txt
View File
| @ -0,0 +1 @@ | |||
| gitpython | |||